YDNA And YDNA Testing
By John Alexander
YDNA Testing: How it all works
You can find all this information in almost any modern book on human biology, but I'll summarize it to make it easy, I hope. Actually, I found it difficult to understand all the discussions of DNA mutation, short tandem repeat (STR), haplotype, allele, single nucleotide, single nucleotide polymorphism (SNP, which is usually just called a SNIP), and haplogroup; therefore, I'll define each term so that anyone interested can look back for reference. Still, I'll try to use most of the terms sparingly, often substituting other words.
To begin, we need to introduce cells, chromosomes, and DNA, all words you probably remember from high-school biology, even if you are as old as I am. Our bodies are formed of cells, and the nucleus of every normal cell in our bodies contains 46 chromosomes, 23 from each parent. Occasionally, there are people with exceptions, but the exceptions are rare. Each chromosome is made up of a DNA molecule wrapped around other material.
Two of these 46 chromosomes are the sex chromosomes. For each female, the sex chromosomes are both X chromosomes, one given by her mother and one given by her father. For each male, one sex chromosome is an X chromosome from his mother, and one is a Y chromosome from his father. The Y chromosome is always inherited from the father; so it comes down to each male from his father, from a grandfather, from a great-grandfather, and on back to a very ancient human male.
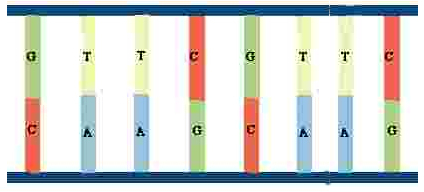
A segment of a DNA molecule might consist of four or five components, perhaps as simple as GTTC (above), repeated over and over with the second G hidden by the helix band. Before the first G, there was some other sequence. Personnel at DNA-testing laboratories and other genetic scientists call the sequence that is repeated a short tandem repeat (STR), but the STRs selected for testing may also be called markers because that is the designation often used by laboratories, and marker is the name we will use. The marker to be tested ends when the sequence changes from repetitions of GATC or whatever sequence was present to repetitions of a different combination, perhaps TAGCC. We need to consider the bases on only one side because when a sequence changes on one side, it changes on the other. This occurs because the same bases are always paired; for example, T as a base on one side of the unwound molecule is always paired with A as the base on the other side. Thanks to mapping done during the Human Genome Project, the exact location on the YDNA molecule, the helix, is known for each marker that genetic laboratories have designated as sites that are suitable for genealogical testing.
As already stated, a segment of DNA, which may be called a marker when it is tested in a laboratory, can have many repetitions of the same sequence of base pairs. The number of repetitions usually ranges from about seven to more than forty; however, the range of values for any given marker is much smaller, with more than ten possible values (alleles, if you wish) being extremey unusual. I will use examples from the YDNA project in which I participated to illustrate the relatively few values that are found, even for markers where mutations occur most frequently. Marker DYS439, a rapidly mutating marker has values ranging from 10 to 14, with almost all the repetitions in our project being 11, 12, or 13. Marker DYS449, also a rapid-mutation site, has values from 26 to 34, but very few at either extreme, mostly 29, 30, 31, or 32. DYS390, which does not mutate rapidly, has values from 21 to 27 but, out of approximately 300 men tested at the time I did the analysis, there were only two values of 21 and only one value of 27. DYS454, which mutates extremely slowly, has only one value, 11, for all members of the project. In the male population at large, values of 10 and 12 occur for marker DYS454, although rarely.
For each person tested, each marker has a specific value, and the collection of those values is his haplotype. People who are closely related should be the same haplotype or have only a few mismatched values; however, there is some possibility of change each time YDNA passes from father to son. Although this means that even a man and his father may have one or more mismatches, the mutation rate is slow enough that mismatches range from zero to four or five (out of sixty-seven tested markers)in the project in which I participated, even for people whose lines split in the days of Colonial American.
In addition to testing for the number of times a sequence of pairs repeats before changing, there is another type of YDNA test that can help in determining relationships, the single nucleotide polymorphism, the SNP or SNIP, mentioned earlier. DNA genealogists and genetic anthropologists define a haplogroup by reference to the SNP that distinguishes it from all other groups. The person in whom the SNP occurred would likely be considered to be in the same haplogroup as his father, uncles, brothers, and cousins, but he might later be assigned the title of a new haplogroup if he has a very large number of male-line descendants, but that would not be known for several generations after he lived.
We don't need to spend much time discusing how SNPs differ from the mutations that change the number of times a sequence repeats because tests for SNPs are seldom needed to distinguish one haplogroup from another. There is not an extremely large number of SNPs in the entire ancestral line of any one man, and all those men in a haplogroup will have the same series of SNPs.
With the necessary words and terms defined simply but not incorrectly, I hope, let us look far, far back in prehistory to the lifetime of the most-recent man from whom all living men descend. This doesn't mean that there were not other men alive at that time also, but, over the centuries and millenia, the lines of the other men ceased having males born to the lines. To make that concept easier to understand, consider that, if you are a male, your great-great grandfather whose surname you bear may have had five brothers and that, out of all six men, he may be the only one with living male-line descendants. Lines die out, as several women seeking male cousins to take the YDNA test have discovered!
Let us call the haplogroup of this ancestor of all living men "A," with the knowledge that there is nothing special about calling it A. We could just as well have called his haplogroup "Jim," if that was the name he used. His father, his brothers and perhaps many uncles and cousins in the neighborhood and surrounding neighborhoods belonged to the same haplogroup and might have objected to their group being dubbed Jim, but that would have been their problem, not ours. Anyhow, the brothers', uncles', and cousins' male lines all died out after many generations, leaving only Jim's male descendants. We have called that descendant and all his descendants haplogroup A, at least up to the point that someone in the group had a SNP. SNIP! Their STR, or marker, values may have differed slightly from one to another, but they all belonged to the same haplogroup.
If Jim's descendant in whom the SNP occurred had a large number of descendants in his own male line, the SNP produced a new haplogroup that we could call A1, which is probably the designation actually given by genetic anthropologists, or we could call the new haplogroup B or X or whatever we wished. Names aren't particularly important except to distinguish one group from another, and the responsible organization changes haplogroup names from time to time. Call the new group A1, and the world now had two patrilineal haplogroups, A and A1. Even if the man in whom the SNP resulted in creation of haplogroup A1 had marker (STR) values almost identical to those of most of his cousins who remained in haplogroup A, marker-changing mutations would occur in both groups as generations passed. Over time, by chance, these marker-changing mutations would likely make the most common marker values in one haplogroup different from the most common marker values in the other.
Since a genetic mutation produced haplogroup A1 from haplogroup A, it shouldn't come as a surprise that additional mutations occurred in A and A1. Some of the mutations were SNPs, and some were increases or decreases in the values associated with DNA markers. Both types occurred, slowly but inevitably, and produced more and more haplogroups and more variation in marker values when comparing members of one haplogroup with members of another.
Over time, each haplogroup has come to have marker values that are almost a signature for that haplogroup, not the value of any specific marker but the values of a group of markers. For example, a genetics expert can look at the marker values for any person in my family that descends from a male born about 1700 and say with almost, but not quite, perfect certainty that the person belongs to the M-222 haplogroup without a test for the M222 SNP being performed. Such discrimination will probably take testing and comparison of at least thirty to forty markers. A lower number may be sufficient to tell that two tested individuals do not have a common ancestor since surname began to be used. The only laboratory in the United States currently doing YDNA testing tests thirty-seven markers, which is probably the minimum one should choose if tested by them.
Using the Alexander Project as an Example of YDNA Testing and Interpretation
What I mention for the Alexander project applies to any surname project that has a sufficient number of men participating in the testing.
Most of the men participating in my surname YDNA project tested at least thirty-seven markers, and and several had sixty-seven markers tested, while a few had tests on one-hundred-eleven markers or more. The genetic scientists at the laboratory performing the tests don't say that two individuals are definitely related but the laboratory provides estimates of the likelihood of the two having a common Alexander ancestor within a given number of generations. The statements they are willing to make can be taken to mean that a common ancestor since around 1500 or 1600 is unlikely if there are mismatches on more than four or five markers out of thirty-seven or more than six or seven out of sixty-seven, the maximum tested by most participants. Based on the laboratory's estimates, several participating Alexanders have so many mismatches with anyone else in the project that it is unlikely they have a common ancestor within the last few thousand years. In some cases, the men have discovered that their male line is not truly Alexander.
In my family project, we have learned that most family groups, as brought together from their YDNA results, have no more than four or five members. This means that there were many origins for the surname Alexander, even within the British Isles, where most of the project participants believe or know their roots to lie. I have looked at other projects and found this to be even more true for more-common surnames, for example, for example, Smith or Taylor.
The family to which almost all United States Alexanders were assigned by early genealogists, who had access to few records and didn't have YDNA-testing to help, is the largest family in our project, but the YDNA mismatches between this family group and my family group, which we can trace to the area around Anson County, NC, and Spartanburg County, SC, suggest we have no common male-line ancestor for at least a thousand years, and the testing laboratory's analysis of a specific mutation pushes the split back to well beyond a thousand years, meaning they and my family are no closer genetically than to almost any person I meet casually on the street.
A few other Alexander families in the project have YDNA profiles fairly similar to my family group, which we whimsically called the Spartanburg Confused group, or the SpartCons. One of these groups with fairly close matches is a family that can trace some of their earliest known ancestors back to Campbelltown, Scotland, and another is a family whose members can mostly trace back to an area in SC near my group's location before dispersing all over the country. Interestingly, this group and the SpartCon group match so closely that there is only a low probability that both groups' Alexander name does not have a common origin. They have also designated their family group as "Confused," the ConsToo or ConsTwo group. Although the laboratory geneticists are hesitant to say that the two groups have a common Alexander ancestor, I have studied the matches and mismatches and have done a statistical analysis that leads me to the conclusion that our common ancestor lived around 1300 to 1500, shortly after the time most Europeans were getting family names, by choice or by assignment. The YDNA profile of the group called the Campbelltown family differs from that of the SpartanCons and the ConsToo much less than is usually found between two families in our project, and the three groups probably have a common ancestor around the year 1200 or a bit earlier. Although it is unlikely that we will ever trace the three families or two of the three to a common ancestor, we can perhaps hope that future advances in DNA analysis will help us trace them to a common time and place.
From the discussion so far, it is likely apparent that, in general, YDNA markers for men of a common surname can match closely or match very poorly, and, if they have tested on 67 markers or more, it is usually easy to determine whether they have a recent common ancestor of that surname. Good matches between individuals with the same surname mean they are likely related unless there is reason to believe otherwise; for example, one man's family has roots in Britain, and the other's family has roots in southern Europe or Russia. Having such close matches may then be due to chance, as it could be for close matches with a person bearing a different surname. After all, probability tells us that, if you continue to flip a coin long enough, you will probably get ten heads or ten tails in a row, although it may take years of flipping to achieve.
Although it is fairly unusual to have an exact match on all tested markers between two people whose most-recent common ancestor lived in the eighteenth century, there is likely to be no more than one, two, or three mismatches even then. Remember, however, that the exactness of the match does not depend directly on the closeness of the kinship. For example, I match exactly on all markers tested, 37 for two of them and 67 for the third, with three Alexanders. I have no common ancestor born later than about 1730 for the two matching on 37 markers and no common ancestor later a generation earlier for the one matching me on 67 markers. In contrast, each of my perfect matches differs on one or more markers with a more-closely related cousin, and I differ by one from a cousin with common ancestry dating from just before 1800. The three of us matching exactly had the most common value on each marker for our family group's testers. None of test participants that we call our family group differed from us three by more than four markers, making all of us almost certain cousins since we can all trace back to a location that is fairly limited in area.
A look at the tables below will show that each of two of the family groups, one we call the Seven Brothers group and one we call the Glasgow group, have so many DNA mismatches to each other and to the SpartCons, the Cons Too, and the Campbelltowns that no paternal-line kinship can exist. The Seven Brothers and the Glasgows are quite representative of the project's family groups in that they are quite distinct from the other families. Very few -- in fact, almost no other -- family groups match as closely as the SpartCons, the Cons Too, and the Campbelltown group match one another. I have not examined closely, but I suspect other surname YDNA projects may have family groups that are fairly close and family groups that differ greatly.
| Family | Haplogroup & Identifying SNP | Mismatches on 67 Sites | Mismatches on 111 Sites |
| SpartCons | R1b1a2a1a1b4b SNP: M222 | -- | -- |
| Seven Brothers | R1b1a2 | 19 | 33 |
| ConsToo | R1b1a2a1a1b4b SNP: M222 | 7 | 14 |
| Campbelltowns | R1b1a2a1a1b4b SNP: M222 | 8 | 16 |
| Glasgows | R1b1a2a1a1b | 23 | 38 |
Just space
| Family | Haplogroup & Identifying SNP | Mismatches on 67 Sites | Mismatches on 111 Sites |
| Seven Brothers | R1b1a2 | - | - |
| SpartCons | R1b1a2a1a1b4b SNP: M222 | 19 | 33 |
| ConsToo | R1b1a2a1a1b4b SNP: M222 | 18 | 30 |
| Campbelltowns | R1b1a2a1a1b4b SNP: M222 | 18 | 33 |
| Glasgows | R1b1a2a1a1b | 16 | 30 |
Just space
| Family | Haplogroup & Identifying SNP | Mismatches on 67 Sites | Mismatches on 111 Sites |
| ConsToo | R1b1a2a1a1b4b SNP: M222 | -- | -- |
| SpartCons | R1b1a2a1a1b4b SNP: M222 | 7 | 14 |
| Seven Brothers | R1b1a2 | 18 | 30 |
| Campbelltowns | R1b1a2a1a1b4b SNP: M222 | 12 | 18 |
| Glasgows | R1b1a2a1a1b | 21 | 34 |
Just space
| Family | Haplogroup & Identifying SNP | Mismatches on 67 Sites | Mismatches on 111 Sites |
| Campbelltowns | R1b1a2a1a1b4b SNP: M222 | -- | -- |
| Seven Brothers | R1b1a2 | 18 | 33 |
| SpartCons | R1b1a2a1a1b4b SNP: M222 | 8 | 16 |
| ConsToo | R1b1a2a1a1b4b SNP: M222 | 12 | 18 |
| Glasgows | R1b1a2a1a1b | 21 | 34 |
Just space
| Family | Haplogroup & Identifying SNP | Mismatches on 67 Sites | Mismatches on 111 Sites |
| Glasgows | R1b1a2a1a1b | -- | -- |
| Seven Brothers | R1b1a2 | 16 | 30 |
| SpartCons | R1b1a2a1a1b4b SNP: M222 | 23 | 38 |
| ConsToo | R1b1a2a1a1b4b SNP: M222 | 21 | 34 |
| Campbelltowns | R1b1a2a1a1b4b SNP: M222 | 21 | 34 |
Conclusions From YDNA Genealogy
We know that our original colonial American ancestor passed on his YDNA to his five sons, four of whom have living descendants who participated in the testing. We know little about the oldest son. All of the four lived in a small area along the NC-SC border. With the YDNA passed on from the father to each of the four sons, the following occurred: (1) most likely, no changes, (2) a small probability of a genetic mutation in one marker, or (3) an extremely low probability of a mutation in more than one of the markers. Each son then passed on YDNA to his sons with zero, one, or more than one change in the series of numbers, and each succeeding generation of males similarly passed on YDNA to sons with some small probability of mutation at each transfer. The YDNA from me, descending from a man we'll call James of Spartanburg, and a test participant descending from James's brother Robert matched exactly on 67 of 67 markers tested, while our DNA and that of a descendant of another brother, David, differed on four markers; however, the DNA of David's descendant matched that of another descendant of James much more closely, meaning that, by chance, the same mutations had occurred.
Sometimes comparison of the values of each marker in a family can reveal additional information. For example, in the line of James of Spartanburg, all tested descendants of one of his sons have a value of 14 on a marker for which all other family members have a value of 13. This tells us a mutation occurred, but we don't know whether it occurred in the transfer from James to the son or from the son to James's grandson, since all this subset of testers descended from that one grandson. We can say only that the defining mutation occurred either when the grandson received the YDNA from his father or when his father received it from James. A test on a descendant of one of that grandson's brothers would be necessary to reveal when the mutation occurred.
Similarly, both tested descendants of another grandson of James of Spartanburg have a value of 29 on a marker for which all other group memebers have a value of 30. These two descendants also have higher values than anyone else on a second marker, and it is of interest to note that they don't have the same value for this second marker, meaning that two separate mutations occurred on this marker for one of them or that one had a two-step mutation.
Mutations such as these that show up on the tests of some members of a family but not other members are often called branch identifiers because they may allow determination of which of several sons in a family was the patriarch of the branch even when there are no records to help.
I will attempt to answer questions emailed to me at jfalex37@comcast.net. It will be posted on the internet at URL http://johnandval.org/genealogy/YDNAGeneral.html, and, if I get a request, I will email a copy of the article to the person requesting it.